Our scoring function consisted of a simple empirical scoring function and a pharmacophore-based scoring function to reduce the number of false positives. The energy function can be dissected into the following terms:
Etot = Ebind + Epharma + Eligpre
where Ebind is the empirical binding energy used during the molecular docking ; Epharma is the energy of binding-site pharmacophores; Eligpre is a penalty value if the ligand unsatisfied the ligand preferences. Epharma and Eligpre were used to improve the number of true positives by discriminating active compounds from hundreds of thousands of non-active compounds. The empirical binding energy (Ebind) is given as
Ebind = Einter + Eintra + Epenal
where Einter and Eintra are the intermolecular and intramolecular energy, respectively, Epenal is a large penalty value if the ligand is out of range of the search box. In this paper, Epenal is set to 10000. The intermolecular energy is defined as
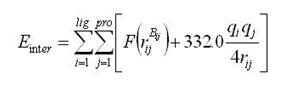
rijBij is the distance between the atoms i and j with the interaction type Bij forming by the pair-wise heavy atoms between ligands and proteins; Bij is eithera hydrogen bond or a steric state; qi and qj are the formal charges and 332.0 is a factor that converts the electrostatic energy into kilocalories per mole. The lig and pro denote the numbers of the heavy atoms in the ligand and receptor, respectively. is a simple atomic pairwise potential function (Figure 1.)
In this atomic pair-wise model, the interactive types are only hydrogen binding and steric potential which have the same function form but with different parameters, V1, . . . , V6 (defined in Figure 1.)
In this model, the atom is divided into four different atom types: donor, acceptor, both, and nonplar. The hydrogen binding can be formed by the following pair atom types: donor-acceptor (or acceptor-donor), donor-both (or both-donor), acceptor-both (or both-acceptor), and both-both. Other pair-atom combinations are to form the steric state.
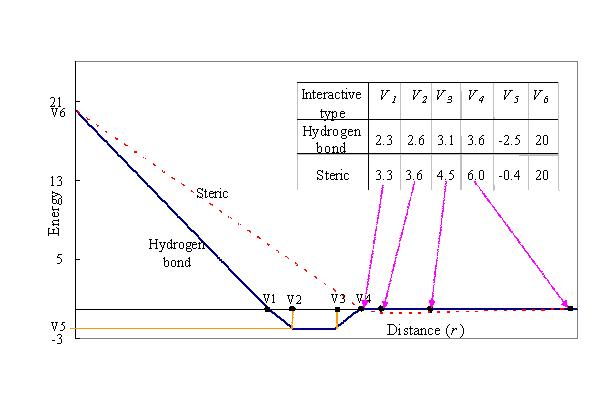
Figure 1. The linear energy function of the pair-wise atoms for the steric interactions (red line) and hydrogen bonds (blue line) in GEMDOCK.
| 