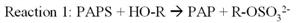
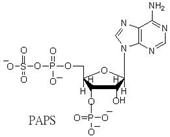
 |
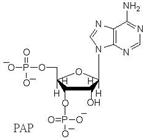 |
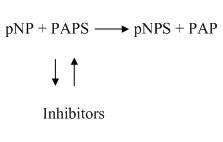
Phenol sulfotransferase (PST) catalyzes sulfuryl group transfer from
adenosine 3'-phosphate 5'-phosphosulfate (PAPS) to a variety of nucleophilic
acceptors in biological system (reaction one). Physiological sulfation by PAPS
results in the production of adenosine 3',5'-diphosphate (PAP). PAP may become a
strong inhibitor or cofactor for the physiological or the transfer reactions,
respectively. Several nucleotides other than PAP also serve as substrate, inhibitor
or cofactor of PST catalyzed reaction (reaction two). We are interested in the effects
of these nucleotides on the PST catalyzed reactions and their possible physiological
significance of in biology. Several nucleotide inhibitors (such as adenosine
5'-diphosphate and adenosine 5'-triphosphate) of sulfotransferase in physiological
reaction have been reported in the literature. However, our data suggests that they
are not inhibitors for the transfer reaction or the reverse physiological reaction
catalyzed by PST. The aim of this work is to show that the molecular docking analysis
can be successfully used to underline the inhibition mechanism of these nucleotides.
Figure 1. The best docked conformations of nucleotides with 1aqu.
We docked all of the tested nucleotides, including PAP, PAPS, AMP, ADP, and ATP, into
the active sites of both the complex of estrogen sulfotransferase with PAP(PDB entry:
1aqu) and the complex of estrogen sulfotransferase with PAPS (PDB entry: 1hy3). Figures
1 and Table 1 show the best docked results. The reference molecules are PAP and PAPS
for 1aqu and 1hy3, respectively, when we calculated the RMSD values for all tested
nucleotides (PAP, PAPS, AMP, ADP, and ATP). According to the RMSD values obtained, these
docked structures for both sulfotransferases can be roughly divided into two categories.
Table 1 shows the RMSD values of PAP, PAPS, AMP, ADP, and ATP that are less than 2.0
angstroms for the sulfotransferase 1aqu and 1hy3. The other category of RMSD values
that are much more than 2.0 angstroms are not given.
| Table 1. The results of nucleotide docking on 1aqu and 1hy3 with GEMDOCK
|
| |
PAP |
PAPS |
AMP |
ADP |
ATP |
1aqu
Minimization Energy of docked structure (cal/mole) |
-192.12 |
-219.96 |
-151.03 |
-154.32 |
-193.83 |
| RMSDa (angstroms) |
0.51 |
1.06 |
1.25 |
0.76 |
1.85 |
1hy3
Minimization Energy of docked structure (cal/mole) |
-192.32 |
-222.97 |
-149.72 |
-173.03 |
-194.60 |
| RMSDb (angstroms) |
1.20 |
0.41 |
1.28 |
1.44 |
1.96 |
aThe values was calculated using reference molecule PAP for these tested nucleotides.
bThe values was calculated using reference molecule PAPS for these tested nucleotides.
|
Several nucleotides (such as PAP, AMP, ADP and ATP) of PST in physiological reaction showed to
inhibit competitively the sulfation of PST when PAPS is the varied substrate (Table 2). Previous
studies showed that various nucleotides bind tightly to PST (Table 3) and some nucleotides, 2',5'-PAP,
AMP, 2':3'-cyclic PAP can be used by PST to replace PAP. Our experimental data suggests that these
nucleotides are not inhibitors of the transfer reaction or the reverse physiological reaction catalyzed
by PST (data not shown). To explain the above experimental data, we report our results on the docking of
nucleotides to EST. These observations are in good agreement with the experimental data reported here.
The docking results explain why all the nucleotides tested bind tightly to PST. It also explains why some
nucleotides may replace PAP as substrates for PST.
| Table 2. Inhibition constants for the competitive inhibition of sulfotransferase by Nucleotidesa |
 |
| Inhibitors |
Ki (mM) |
| PAP |
0.1 (0.01)b |
| AMP |
>1000 |
| ADP |
30.0 (1.2) |
| ATP |
23.2 (0.7) |
a Data obtained from Rens-Domiano, S. S.; Roth, J. A. J Neurochem. 1987, 48, 1411-1415
b Deviation between +0.01 and -0.01 |
| Table 3. Nucleotides used for the docking in PST |
| Name |
Dissociation constants |
Abbrevation |
| |
Kd1 (nM) |
Kd2 (microM) |
|
| 3'-phosphoadenosine 5'-phosphate |
31 (4)a, b
20 c |
152 (1)b
200 c |
PAP |
| 3'-phosphoadenosine 5'-phosphousulfate |
>>c |
>>c |
PAPS |
| Adenosine 5'-monophosphate |
9.2 (0.3) |
164 (4) |
AMP |
| Adenosine 5'-diphosphate |
38 (1) |
75.0 (0.3) |
ADP |
| Adenosine 5'-triphosphate |
79 (1) |
122 (1) |
ATP |
a Deviation between +4 and -4
b Data obtained from Lin, E. S.; Yang, Y. S. Biochem Biophys Res Commun. 2000, 271, 818-822
c Data was too large to be measured. (Yang, Y. S.; Marshall, A. D.; McPhie, P.; Guo, W. X.; Xie, X.; Chen, X.; Jakoby, W. B. Protein Expression and Purification 1996, 8, 423-429)
|
Through the use of RMSD values, one can predict to which enzyme a specific substrate/inhibitor will best bind. In this study, it was known from experimental data that PAP binds better than other nucleotide to sulfotransferase.
|
