Introduction |
| What can KIDFamMap do for you? |
|
KIDFamMap is the first database grouping 189,987 kinase-inhibitor interactions (from BindingDB and kinase profiling) into 1,210 pharma-interfaces (called kinase-inhibitor family), deriving the relationship between the moiety preferences and physico-chemical properties of binding sites of kinases and inhibitors, relationships between 399 human protein kinase (from Kinase.Com), 35,788 kinase inhibitors (from BindingDB and kinase profiling) and 339 diseases (from OMIM and KEGG). KIDFamMap groups kinase-inhibitor interactions into different families based on their pharma-interfaces. For each family, KIDFamMap shows the pharma-interface and interaction between kinase and selected inhibitor. KIDFamMap identifies moiety preferences and physico-chemical properties of binding sites of selected kinase, anchors and inhibitor-anchor map. |
| How to use KIDFamMap database |
|
| FASTA Format |
|
Each query sequences is in FASTA format consists of a single-line description, followed by lines of sequence data. The first character of the description line is a ">" symbol in the first column. >ABL1 GMSPNYDKWEMERTDITMKHKLGGGQYGEVYEGVWKKYSLTVAVKTLKEDTMEVEEFLKE AAVMKEIKHPNLVQLLGVCTREPPFYIITEFMTYGNLLDYLRECNRQEVNAVVLLYMATQ ISSAMEYLEKKNFIHRDLAARNCLVGENHLVKVADFGLSRLMTGDTYTAPAGAKFPIKWT APESLAYNKFSIKSDVWAFGVLLWEIATYGMSPYPGIDLSQVYELLEKDYRMERPEGCPE KVYELMRACWQWNPSDRPSFAEIHQAFETMFQESSISDEVEKELGKQ |
Explanation of outputs |
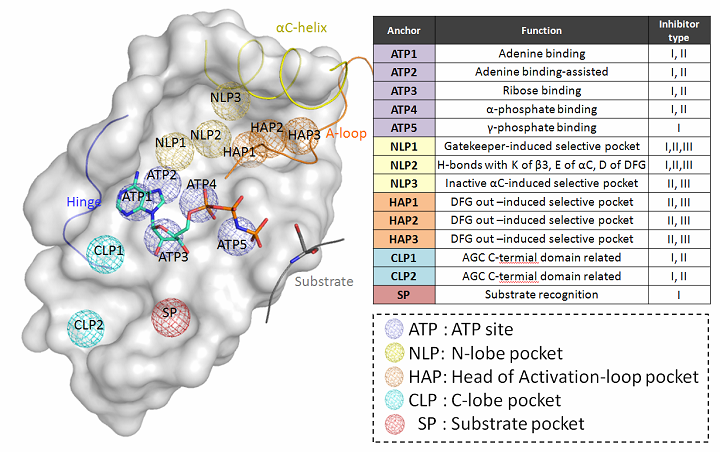
| Explanation of KIDFamMap anchors |
|
|
| Explanation of kinase comformation types |
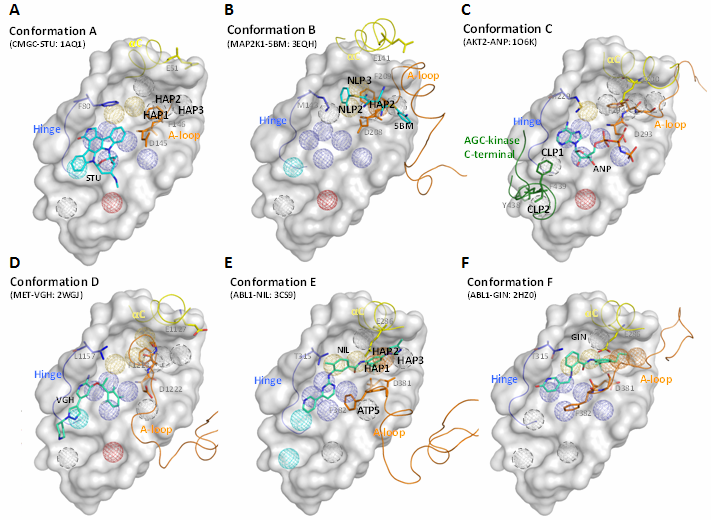
Kinase conformations are highly correlated to catalytic activity and inhibitor selectivity (kinase inhibitor types). Based on 14 KIDFamMap anchors, we can divide kinase conformations into six types:
|
| Kinase related diseases |
|
Protein kinases are one of the most important classes of drug targets because the deregulation of kinase functions is often implicated in many diseases such as cancers, neurological and metabolic diseases. These kinase related diseases are collected from OMIM and KEGG. KIDFamMap provides 962 kinase-inhibitor-disease relationships and 339 diseases. |
| Allelic variants |
|
The allelic variants are collected from OMIM. There are 638 disease allelic variants in KIDFamMap. Most of the allelic variants represent disease-causing mutations. A few polymorphisms are included, many of which show a positive correlation with particular common disorders. |
| Pocket pattern |
|
For each identified anchors in the kinase-inhibitor familes, KIDFamMap provides the conservations of identified anchors with Weblogo. These pocket patterns presents the sequence conservations of surroundings of interacting residues. |
| 3D presentation of KIDFamMap |
|
The result will be shown as data tables and visualized in 3D presentation. Jmol applet is embedded for 3D presentation. We suggest users' browser to install Java Runtime Environment for running Jmol applet. |
| Presentation of compound structures |
|
We present compound structures by using OASA library which is a python library for manipulation of chemical formats that forms the base of BKChem. |